| Manufactured By: | Sino Biological Inc. |
| Synonym: | SuperNuclease, Nuclease, Serratia marcescens' extracellular endonuclease, Benzonase®; equivalent (Benzonase®; endonuclease is a trademark of Merck KGaA, Darmstadt, Germany). |
| Protein Construction: | Construction of SuperNuclease is similar to its natural form. |
| Source: | Serratia marcescens |
| Expression Host: | Escherichia coli (E. coli). |
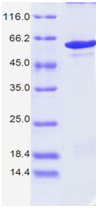
| Purity: | > 98%, as determined by SDS-PAGE |
| Endotoxin: | Less than 0.22 EU/mg of the SuperNuclease (SSNP01) as determined by the LAL method. |
| Stability: | Samples are stable for up to twelve months from date of receipt -70℃. |
| Predicted N terminal: | Three isoforms with different N terminal may be found from the compound—Sm1 (22D-266N), Sm2 (23T-266N) and Sm3 (25E-266N), the activity analysis shows that they were functionally equivalent[6, 15]. |
|
| Fig. 1. |
SDS-PAGE:
|
| Fig. 2. |
| Shipment: | Shipped at ambient temperature. The liquid or lyophilized enzyme is stable for at least 21d when stored at 37 ℃ (or ambient temperature). |
| Formulation | SuperNuclease is lyophilized from sterile 50 mM Tris-HCl pH 8.0, 20 mM NaCl, 2 mM MgCl2, 5 % trehalose, 5 % mannitol, and 0.01 % Triton® (Triton® is a trademark of Union Carbide, USA). Follow the instructions on the vial. Centrifuge the vial at 4℃ before opening to recover the entire contents. Please contact us for any concerns or special requirements. |
| Reconstitution: | Follow the instructions on the vial. Centrifuge the vial at 4℃ before opening to recover the entire contents. Normaly,25U/μL is recommended to be the final concentration. |
| Storage: | Store it under sterile conditions at -20℃ to -80℃ upon receiving for at least 12 months. Recommend to aliquot the protein into smaller quantities for optimal storage. Avoid repeated freeze-thaw cycles. |
(1).Viscosity Reduction of E. coli Lysate
Cell extracts often show high viscosity due to nucleic acids. The viscosity of lysate may lead to reduction of the protein production. This problem can be solved by using SuperNuclease.
Experimental design
E. coli (1.0 g) with a recombined pET-28a construct was suspended in cell lysate buffer (50 mM Tris-HCl (pH 8.0), 4 M Urea, 100 mM DTT, and 1% Triton X-100), and resulted in 1.0 g/mL. Then cell lysate was incubated with SuperNuclease at 4℃ for 5 min. Then samples were centrifuged at 10000 g for 1 min, and photographed.
Result
|
| Fig. 3. A. With 25U/mL SuperNuclease |
(2).Nucleic Acid Elimination of Nucleoprotein (NP)
Nucleoprotein of virus is always thought to be an attractive target for vaccine development. Biotech has make it possible to express and purify the NP from the engineered E. coli, Baculovirus-insect cells or other expression hosts.
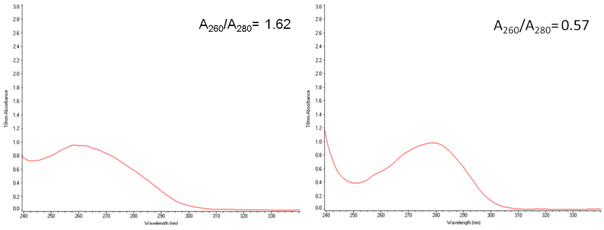
However, reducing of the nucleic acid contamination is needed for successful development of the vaccine. This problem can be solved by using SuperNuclease, which can decrease the viscosity of the cell lysate, reducing the ratio of A260/A280.
Experimental design
Expression of NP took place over 48 h at 26℃, cell plate was collected by centrifugation, and washed with buffer A (20 mM Tris pH8.0, 50 mM NaCl, 5 mM MgCl2). Cells were lysed by ultrasonic probe in buffer A (25 U/mL SuperNuclease). Supernatant contained NP-(His tag) was collected by centrifugation for 20 min, at 10000 g, 4℃, then NP was adhered to Ni+ column, and washed with low concentration of imidazole, high concentration of NaCl (to separate the fragment of nucleic acid from protein), low concentration of NaCl, and proper concentration of imidazole to elute the protein from the column. Finally, NP was desalted and stored in the proper buffer.
Result
 |  |
| A | B |
 | |
| C | D |
| Fig. 4. SDS-PAGE (15%) gel electrophoresis of Nucleoprotein, and it's A260/A280=0.57 A and C: The sample without SuperNuclease treatment; B and D. The sample with SuperNuclease treatment. | |
(3).Salmon sperm DNA and plasmid DNA cleavage assay
The cleavage activity unit is defined as the amount of SuperNuclease that degrades DNA and causes 1 Absorbance Unit change at 260 nm (37℃ for 30 minutes) which corresponds to complete digestion of 37 µg of DNA.
The phenomenon (nucleic acid digestion) can be shown by agarose gel electrophoresis. Here we use two kinds of DNA as the substrate to perform this reaction.
Experimental design
The substrate Deoxyribonucleic acid sodium salt from salmon testes (Sigma, Catalog # D1626) and plasmid DNA pCMV-Fc-His (Sino Biological Inc. Catalog # CV008) are diluted with assay buffer (50 mM Tris-HCl pH 8.0, 5 mM MgCl2, 100 μg/mL bovine serum albumin) into 1 mg/mL. Incubate the substrate with different units of SuperNuclease as well as other nucleases at 37℃ for 30 min. The DNA fragment is analyzed by agarose gel electrophoresis, and photographed.
Result
|
| Fig. 5. |
|
| Fig. 6. |
The SuperNuclease is a nonspecific nuclease with high activity, capable of completely digesting RNA and DNA (single stranded, double stranded, linear, circular and super coiled forms, that no fewer than five phosphate residues [1]) into 5'-monophosphate-terminated oligonucleotides of 3-5 bases in length [2]. SuperNuclease requires divalent cation, preferably Mg2+ for activity, displays a broad pH tolerance (range from 6 to 10, optimal at 8-8.5) and has a wide temperature optimum between 35℃ and 44℃ [3]. The nuclease is a homodimer (the dimer form is physiologic and functions more progressively than the monomer [3,4,5,6,7]). Two disulfide bonds in the nuclease are crucial to its activity and stability [9]. It does not have typical protease activity detected by azocasein assay. Its high intrinsic activity and broad substrate tolerance make the endonuclease an ideal tool in a variety of biotechnological and pharmaceutical applications. SuperNuclease can be removed by various purification methods.
温馨提示:因厂家更改产品包装、产地或者更换随机附件等没有任何提前通知,且每位咨询者购买情况、提问时间等不同,为此以下回复仅对提问者3天内有效,其他网友仅供参考!若由此给您带来不便请多多谅解,谢谢!
服务热线
0771-3293894